Latest Posts
- By Matt GaidicaApril 26, 2023
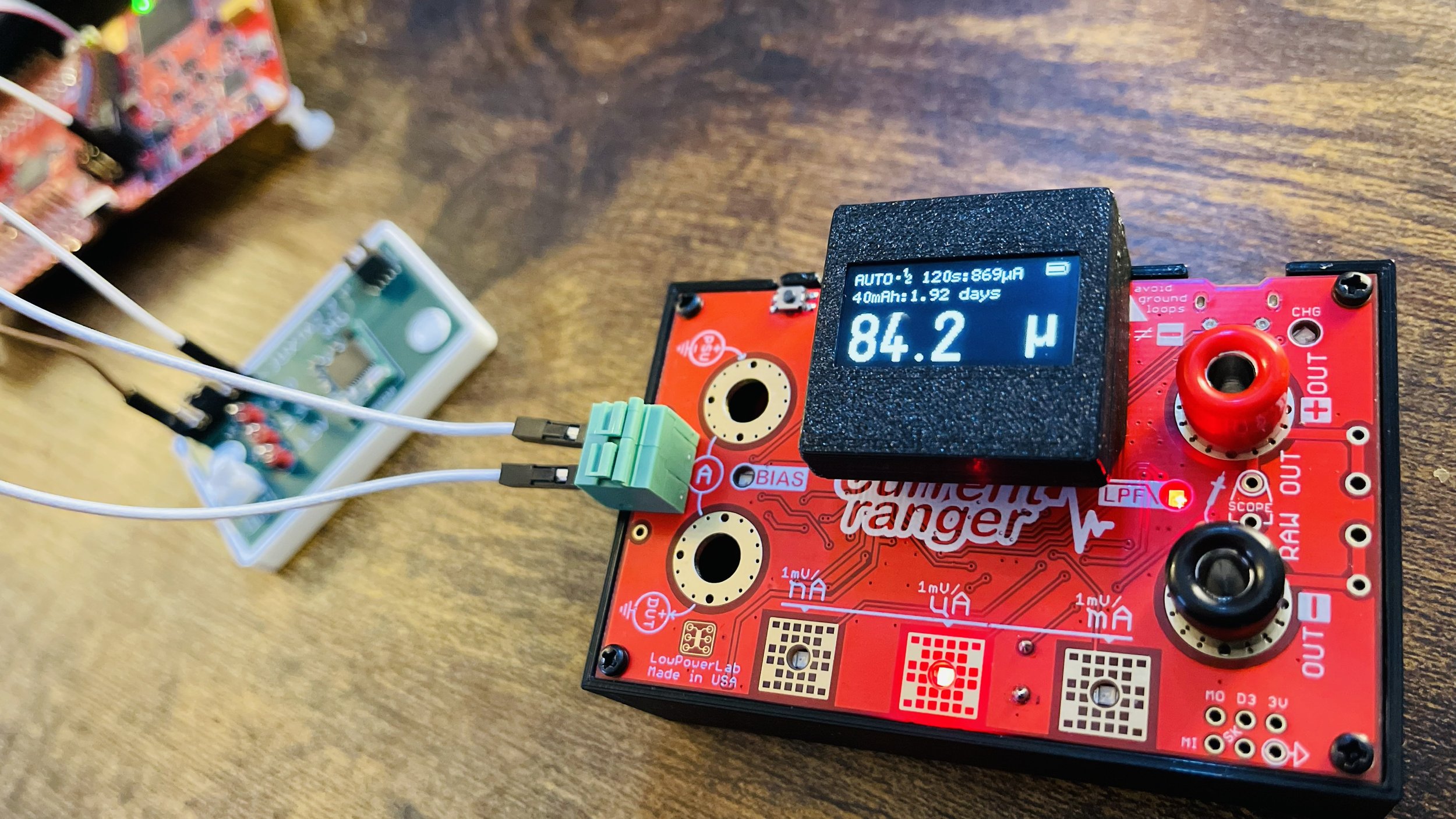
I came upon Current Ranger from LowPowerLab and have been amazed at how feature-full it is, not to mention the ability to
- By Matt GaidicaApril 11, 2023
Watto for iOS A play on watts, the standard unit of power. Watto makes power profiling at the µ-scale easier than before.
- By Matt GaidicaJanuary 31, 2023
A picture is worth 1,000 words. And this is why images are so useful in science communication. The right image only needs
- By Matt GaidicaNovember 9, 2022
GR/PR on iOS In animal models of limb disuse, grip strength (or force) is a measure used to characterize the time course
- By Matt GaidicaMay 19, 2022
Given the important connection between the peripheral and central nervous systems, heart and respiratory rates have become valuable measures to incorporate into
- By Matt GaidicaMarch 14, 2022
Minimums for visual flight rules (VFR) airspace has always been tricky for me to remember. Popular opinion has Rod Machado’s technique at
- By Matt GaidicaJanuary 26, 2022
I can’t tell you how many times I have shared a figure from MATLAB using beta-level code and then forgot what file
- By Matt GaidicaDecember 28, 2021
Hindlimb Unloading Model of Microgravity (see Gnyubkin and Vico, 2015). Load on Forelimbs (left, black) and Tension on Tail (right, red) versus
- By Matt GaidicaApril 27, 2021
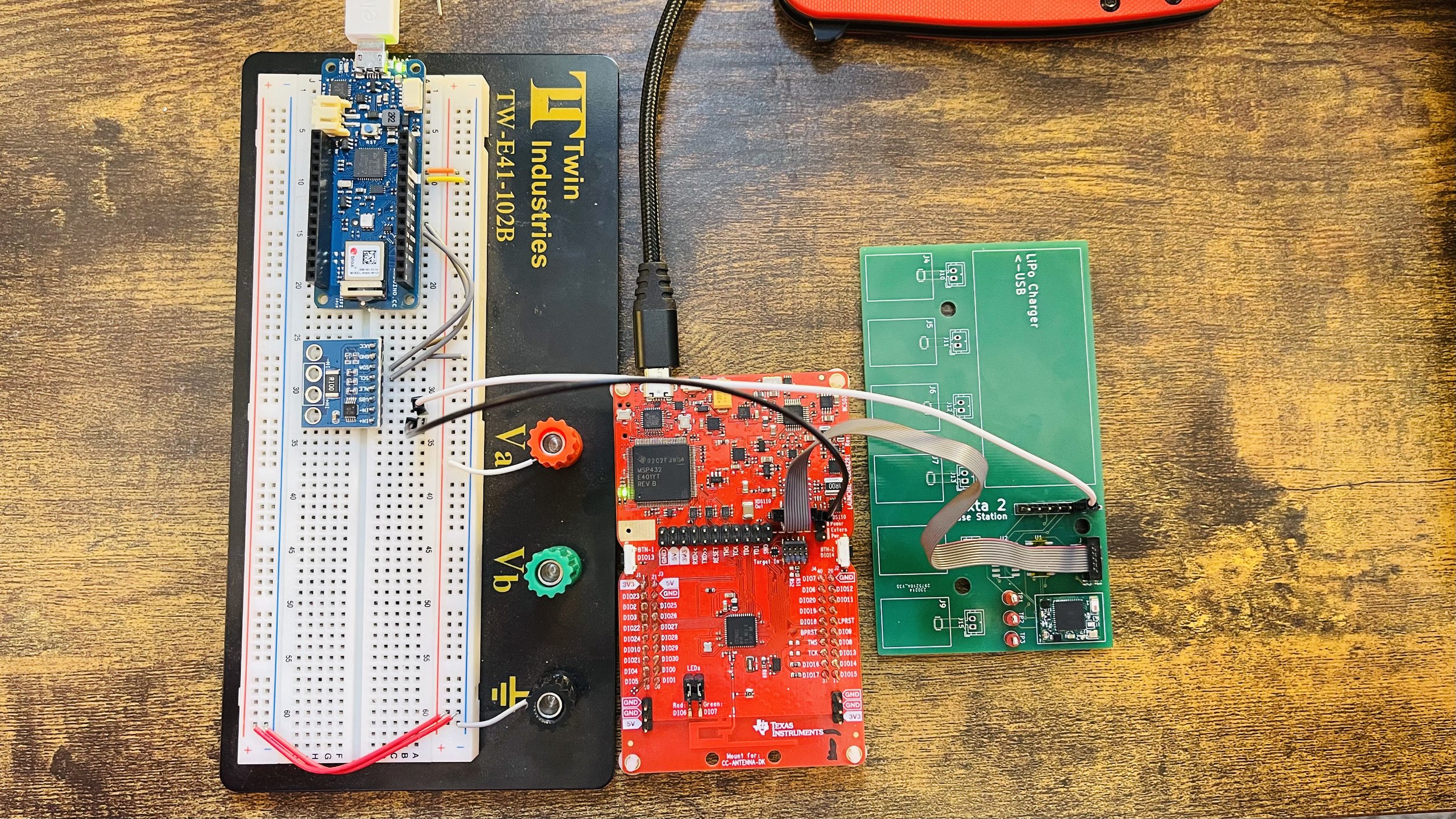
BLE-enabled bio-loggers make it possible to monitor data and change settings on-the-fly. “ESLO” is our custom bio-logger device that pairs with this
- By Matt GaidicaDecember 16, 2020
Zero-padding a Fast Fourier Transform (FFT) can increase the resolution of the frequency domain results (see FFT Zero Padding). This is useful